[1]:
%matplotlib inline
Comparison of machine learning algorithms
[1]:
from ai4water.datasets import busan_beach
from ai4water.utils.utils import get_version_info
from ai4water.experiments import MLRegressionExperiments
for k,v in get_version_info().items():
print(f"{k} version: {v}")
python version: 3.8.13 | packaged by conda-forge | (default, Mar 25 2022, 05:59:00) [MSC v.1929 64 bit (AMD64)]
os version: nt
ai4water version: 1.06
tcn version: 3.5.0
xgboost version: 1.7.0
easy_mpl version: 0.21.3
SeqMetrics version: 1.3.3
tensorflow version: 2.7.0
keras.api._v2.keras version: 2.7.0
numpy version: 1.22.2
pandas version: 1.5.1
matplotlib version: 3.6.1
h5py version: 3.6.0
sklearn version: 1.1.3
xarray version: 0.21.1
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\experimental\enable_hist_gradient_boosting.py:16: UserWarning: Since version 1.0, it is not needed to import enable_hist_gradient_boosting anymore. HistGradientBoostingClassifier and HistGradientBoostingRegressor are now stable and can be normally imported from sklearn.ensemble.
warnings.warn(
[2]:
data = busan_beach()
data.head()
[2]:
| tide_cm | wat_temp_c | sal_psu | air_temp_c | pcp_mm | pcp3_mm | pcp6_mm | pcp12_mm | wind_dir_deg | wind_speed_mps | air_p_hpa | mslp_hpa | rel_hum | tetx_coppml | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| index | ||||||||||||||
| 2018-06-19 00:00:00 | 36.407149 | 19.321232 | 33.956058 | 19.780000 | 0.0 | 0.0 | 0.0 | 0.0 | 159.533333 | 0.960000 | 1002.856667 | 1007.256667 | 95.000000 | NaN |
| 2018-06-19 00:30:00 | 35.562515 | 19.320124 | 33.950508 | 19.093333 | 0.0 | 0.0 | 0.0 | 0.0 | 86.596667 | 0.163333 | 1002.300000 | 1006.700000 | 95.000000 | NaN |
| 2018-06-19 01:00:00 | 34.808016 | 19.319666 | 33.942532 | 18.733333 | 0.0 | 0.0 | 0.0 | 0.0 | 2.260000 | 0.080000 | 1001.973333 | 1006.373333 | 95.000000 | NaN |
| 2018-06-19 01:30:00 | 30.645216 | 19.320406 | 33.931263 | 18.760000 | 0.0 | 0.0 | 0.0 | 0.0 | 62.710000 | 0.193333 | 1001.776667 | 1006.120000 | 95.006667 | NaN |
| 2018-06-19 02:00:00 | 26.608980 | 19.326729 | 33.917961 | 18.633333 | 0.0 | 0.0 | 0.0 | 0.0 | 63.446667 | 0.510000 | 1001.743333 | 1006.103333 | 95.006667 | NaN |
[3]:
print(data.shape)
(1446, 14)
[4]:
comparisons = MLRegressionExperiments(
input_features=data.columns.tolist()[0:-1],
output_features=data.columns.tolist()[-1:],
split_random=True,
verbosity=0,
save=False,
)
[5]:
comparisons.fit(data=data,
run_type="dry_run")
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
running ARDRegression model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
running AdaBoostRegressor model
running BaggingRegressor model
running BayesianRidge model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\tree\_classes.py:306: FutureWarning: `max_features='auto'` has been deprecated in 1.1 and will be removed in 1.3. To keep the past behaviour, explicitly set `max_features=1.0'`.
warnings.warn(
running DecisionTreeRegressor model
running DummyRegressor model
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_coordinate_descent.py:648: ConvergenceWarning: Objective did not converge. You might want to increase the number of iterations, check the scale of the features or consider increasing regularisation. Duality gap: 7.472e+14, tolerance: 2.710e+11
model = cd_fast.enet_coordinate_descent(
running ElasticNet model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
running ElasticNetCV model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\tree\_classes.py:306: FutureWarning: `max_features='auto'` has been deprecated in 1.1 and will be removed in 1.3. To keep the past behaviour, explicitly set `max_features=1.0'`.
warnings.warn(
running ExtraTreeRegressor model
running ExtraTreesRegressor model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\ensemble\_forest.py:416: FutureWarning: `max_features='auto'` has been deprecated in 1.1 and will be removed in 1.3. To keep the past behaviour, explicitly set `max_features=1.0` or remove this parameter as it is also the default value for RandomForestRegressors and ExtraTreesRegressors.
warn(
running GaussianProcessRegressor model
running GradientBoostingRegressor model
running HistGradientBoostingRegressor model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_huber.py:335: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
self.n_iter_ = _check_optimize_result("lbfgs", opt_res, self.max_iter)
running HuberRegressor model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running KNeighborsRegressor model
running KernelRidge model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_ridge.py:251: UserWarning: Singular matrix in solving dual problem. Using least-squares solution instead.
warnings.warn(
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_base.py:133: FutureWarning: The default of 'normalize' will be set to False in version 1.2 and deprecated in version 1.4.
If you wish to scale the data, use Pipeline with a StandardScaler in a preprocessing stage. To reproduce the previous behavior:
from sklearn.pipeline import make_pipeline
model = make_pipeline(StandardScaler(with_mean=False), Lars())
If you wish to pass a sample_weight parameter, you need to pass it as a fit parameter to each step of the pipeline as follows:
kwargs = {s[0] + '__sample_weight': sample_weight for s in model.steps}
model.fit(X, y, **kwargs)
warnings.warn(
running Lars model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_base.py:133: FutureWarning: The default of 'normalize' will be set to False in version 1.2 and deprecated in version 1.4.
If you wish to scale the data, use Pipeline with a StandardScaler in a preprocessing stage. To reproduce the previous behavior:
from sklearn.pipeline import make_pipeline
model = make_pipeline(StandardScaler(with_mean=False), LarsCV())
If you wish to pass a sample_weight parameter, you need to pass it as a fit parameter to each step of the pipeline as follows:
kwargs = {s[0] + '__sample_weight': sample_weight for s in model.steps}
model.fit(X, y, **kwargs)
warnings.warn(
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
running LarsCV model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_coordinate_descent.py:648: ConvergenceWarning: Objective did not converge. You might want to increase the number of iterations, check the scale of the features or consider increasing regularisation. Duality gap: 9.650e+14, tolerance: 2.710e+10
model = cd_fast.enet_coordinate_descent(
running Lasso model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
running LassoCV model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_base.py:133: FutureWarning: The default of 'normalize' will be set to False in version 1.2 and deprecated in version 1.4.
If you wish to scale the data, use Pipeline with a StandardScaler in a preprocessing stage. To reproduce the previous behavior:
from sklearn.pipeline import make_pipeline
model = make_pipeline(StandardScaler(with_mean=False), LassoLars())
If you wish to pass a sample_weight parameter, you need to pass it as a fit parameter to each step of the pipeline as follows:
kwargs = {s[0] + '__sample_weight': sample_weight for s in model.steps}
model.fit(X, y, **kwargs)
Set parameter alpha to: original_alpha * np.sqrt(n_samples).
warnings.warn(
running LassoLars model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_base.py:133: FutureWarning: The default of 'normalize' will be set to False in version 1.2 and deprecated in version 1.4.
If you wish to scale the data, use Pipeline with a StandardScaler in a preprocessing stage. To reproduce the previous behavior:
from sklearn.pipeline import make_pipeline
model = make_pipeline(StandardScaler(with_mean=False), LassoLarsCV())
If you wish to pass a sample_weight parameter, you need to pass it as a fit parameter to each step of the pipeline as follows:
kwargs = {s[0] + '__sample_weight': sample_weight for s in model.steps}
model.fit(X, y, **kwargs)
Set parameter alpha to: original_alpha * np.sqrt(n_samples).
warnings.warn(
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
running LassoLarsCV model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\SeqMetrics\_rgr.py:1148: RuntimeWarning: invalid value encountered in true_divide
zy = (self.predicted - np.mean(self.predicted)) / np.std(self.predicted, ddof=1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_base.py:133: FutureWarning: The default of 'normalize' will be set to False in version 1.2 and deprecated in version 1.4.
If you wish to scale the data, use Pipeline with a StandardScaler in a preprocessing stage. To reproduce the previous behavior:
from sklearn.pipeline import make_pipeline
model = make_pipeline(StandardScaler(with_mean=False), LassoLarsIC())
If you wish to pass a sample_weight parameter, you need to pass it as a fit parameter to each step of the pipeline as follows:
kwargs = {s[0] + '__sample_weight': sample_weight for s in model.steps}
model.fit(X, y, **kwargs)
Set parameter alpha to: original_alpha * np.sqrt(n_samples).
warnings.warn(
running LassoLarsIC model
running LinearRegression model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running LinearSVR model
running MLPRegressor model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\neural_network\_multilayer_perceptron.py:702: ConvergenceWarning: Stochastic Optimizer: Maximum iterations (200) reached and the optimization hasn't converged yet.
warnings.warn(
running NuSVR model
running OneClassSVM model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_base.py:133: FutureWarning: The default of 'normalize' will be set to False in version 1.2 and deprecated in version 1.4.
If you wish to scale the data, use Pipeline with a StandardScaler in a preprocessing stage. To reproduce the previous behavior:
from sklearn.pipeline import make_pipeline
model = make_pipeline(StandardScaler(with_mean=False), OrthogonalMatchingPursuit())
If you wish to pass a sample_weight parameter, you need to pass it as a fit parameter to each step of the pipeline as follows:
kwargs = {s[0] + '__sample_weight': sample_weight for s in model.steps}
model.fit(X, y, **kwargs)
warnings.warn(
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_omp.py:757: RuntimeWarning: Orthogonal matching pursuit ended prematurely due to linear dependence in the dictionary. The requested precision might not have been met.
coef_, self.n_iter_ = orthogonal_mp_gram(
running OrthogonalMatchingPursuit model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_base.py:133: FutureWarning: The default of 'normalize' will be set to False in version 1.2 and deprecated in version 1.4.
If you wish to scale the data, use Pipeline with a StandardScaler in a preprocessing stage. To reproduce the previous behavior:
from sklearn.pipeline import make_pipeline
model = make_pipeline(StandardScaler(with_mean=False), OrthogonalMatchingPursuitCV())
If you wish to pass a sample_weight parameter, you need to pass it as a fit parameter to each step of the pipeline as follows:
kwargs = {s[0] + '__sample_weight': sample_weight for s in model.steps}
model.fit(X, y, **kwargs)
warnings.warn(
running OrthogonalMatchingPursuitCV model
running PoissonRegressor model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_glm\glm.py:294: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
self.n_iter_ = _check_optimize_result("lbfgs", opt_res)
running RANSACRegressor model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running RadiusNeighborsRegressor model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\neighbors\_regression.py:482: UserWarning: One or more samples have no neighbors within specified radius; predicting NaN.
warnings.warn(empty_warning_msg)
D:\mytools\AI4Water\ai4water\experiments\_main.py:2262: UserWarning: model RadiusNeighborsRegressor predicted only nans
warnings.warn(f"model {model.model_name} predicted only nans")
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\ensemble\_forest.py:416: FutureWarning: `max_features='auto'` has been deprecated in 1.1 and will be removed in 1.3. To keep the past behaviour, explicitly set `max_features=1.0` or remove this parameter as it is also the default value for RandomForestRegressors and ExtraTreesRegressors.
warn(
running RandomForestRegressor model
running Ridge model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running RidgeCV model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running SGDRegressor model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running SVR model
running TheilsenRegressor model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_theil_sen.py:128: ConvergenceWarning: Maximum number of iterations 50 reached in spatial median for TheilSen regressor.
warnings.warn(
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\linear_model\_glm\glm.py:294: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
self.n_iter_ = _check_optimize_result("lbfgs", opt_res)
running TweedieRegressor model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running XGBRFRegressor model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\mytools\AI4Water\ai4water\utils\utils.py:1209: RuntimeWarning: Precision loss occurred in moment calculation due to catastrophic cancellation. This occurs when the data are nearly identical. Results may be unreliable.
stats[feat] = np.round(point_features[feat](data), precision)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
running XGBRegressor model
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\neighbors\_regression.py:482: UserWarning: One or more samples have no neighbors within specified radius; predicting NaN.
warnings.warn(empty_warning_msg)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
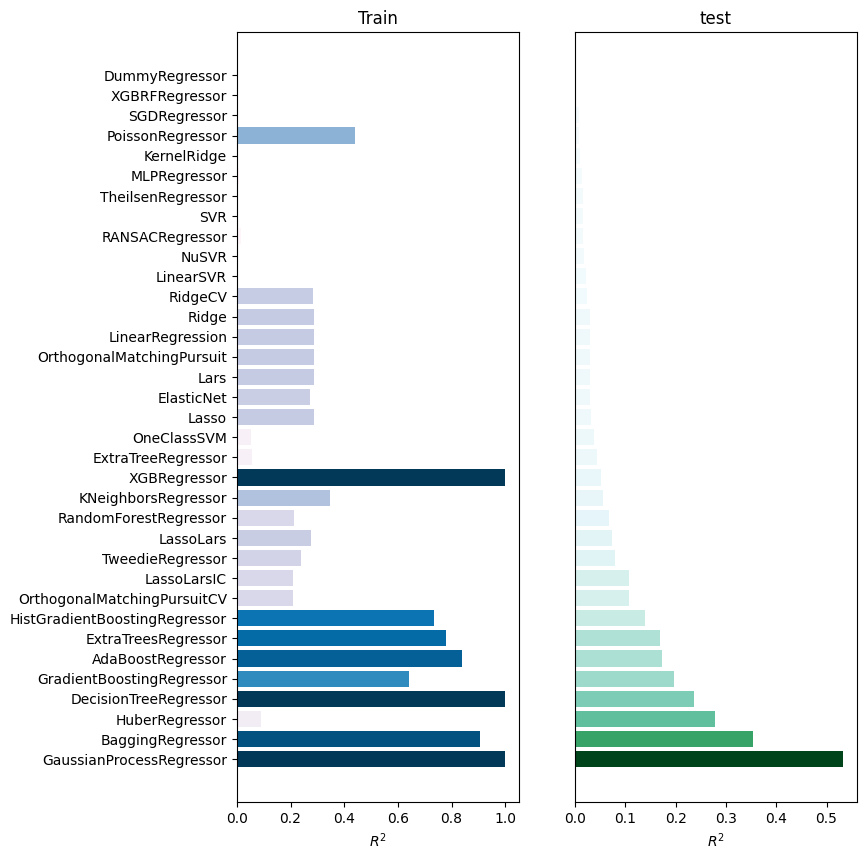
train test
GaussianProcessRegressor 1.000000e+00 5.335382e-01
BaggingRegressor 9.043672e-01 3.529782e-01
HuberRegressor 8.881048e-02 2.784062e-01
DecisionTreeRegressor 1.000000e+00 2.371542e-01
GradientBoostingRegressor 6.428487e-01 1.957935e-01
AdaBoostRegressor 8.395908e-01 1.730707e-01
ExtraTreesRegressor 7.778136e-01 1.693949e-01
HistGradientBoostingRegressor 7.337992e-01 1.385193e-01
OrthogonalMatchingPursuitCV 2.074816e-01 1.076211e-01
LassoLarsIC 2.074816e-01 1.076211e-01
TweedieRegressor 2.396319e-01 7.946911e-02
LassoLars 2.745042e-01 7.419891e-02
RandomForestRegressor 2.112886e-01 6.679858e-02
KNeighborsRegressor 3.470804e-01 5.621710e-02
XGBRegressor 1.000000e+00 5.250909e-02
ExtraTreeRegressor 5.824179e-02 4.286954e-02
OneClassSVM 5.115284e-02 3.817905e-02
Lasso 2.872411e-01 3.169839e-02
ElasticNet 2.729841e-01 3.059856e-02
Lars 2.875785e-01 3.026891e-02
OrthogonalMatchingPursuit 2.875817e-01 3.026343e-02
LinearRegression 2.875798e-01 3.026041e-02
Ridge 2.868481e-01 2.924577e-02
RidgeCV 2.849961e-01 2.390976e-02
LinearSVR 2.083020e-03 2.208388e-02
NuSVR 1.582407e-03 1.717038e-02
RANSACRegressor 1.548905e-02 1.648130e-02
SVR 1.752816e-03 1.620288e-02
TheilsenRegressor 4.240463e-03 1.606258e-02
MLPRegressor 6.478158e-03 1.397927e-02
KernelRidge 3.736431e-03 1.070258e-02
PoissonRegressor 4.394489e-01 7.535408e-03
SGDRegressor 9.470476e-04 7.331519e-03
XGBRFRegressor 8.559689e-35 1.067836e-32
DummyRegressor 6.933348e-35 6.582350e-33
[8]:
best_models = comparisons.compare_errors(
'r2',
data=data,
cutoff_type='greater',
cutoff_val=0.1,
figsize=(8, 9)
)
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\neighbors\_regression.py:482: UserWarning: One or more samples have no neighbors within specified radius; predicting NaN.
warnings.warn(empty_warning_msg)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
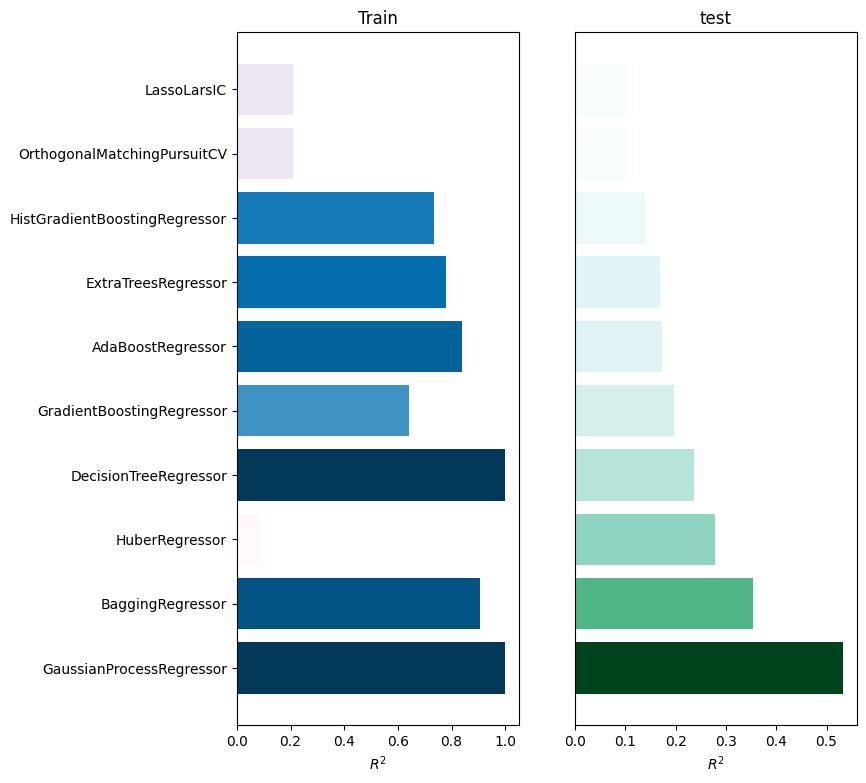
[9]:
train test
GaussianProcessRegressor 1.000000 0.533538
BaggingRegressor 0.904367 0.352978
HuberRegressor 0.088810 0.278406
DecisionTreeRegressor 1.000000 0.237154
GradientBoostingRegressor 0.642849 0.195794
AdaBoostRegressor 0.839591 0.173071
ExtraTreesRegressor 0.777814 0.169395
HistGradientBoostingRegressor 0.733799 0.138519
OrthogonalMatchingPursuitCV 0.207482 0.107621
LassoLarsIC 0.207482 0.107621
[10]:
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\neighbors\_regression.py:482: UserWarning: One or more samples have no neighbors within specified radius; predicting NaN.
warnings.warn(empty_warning_msg)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2829: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[:, None]
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\numpy\lib\function_base.py:2830: RuntimeWarning: invalid value encountered in true_divide
c /= stddev[None, :]
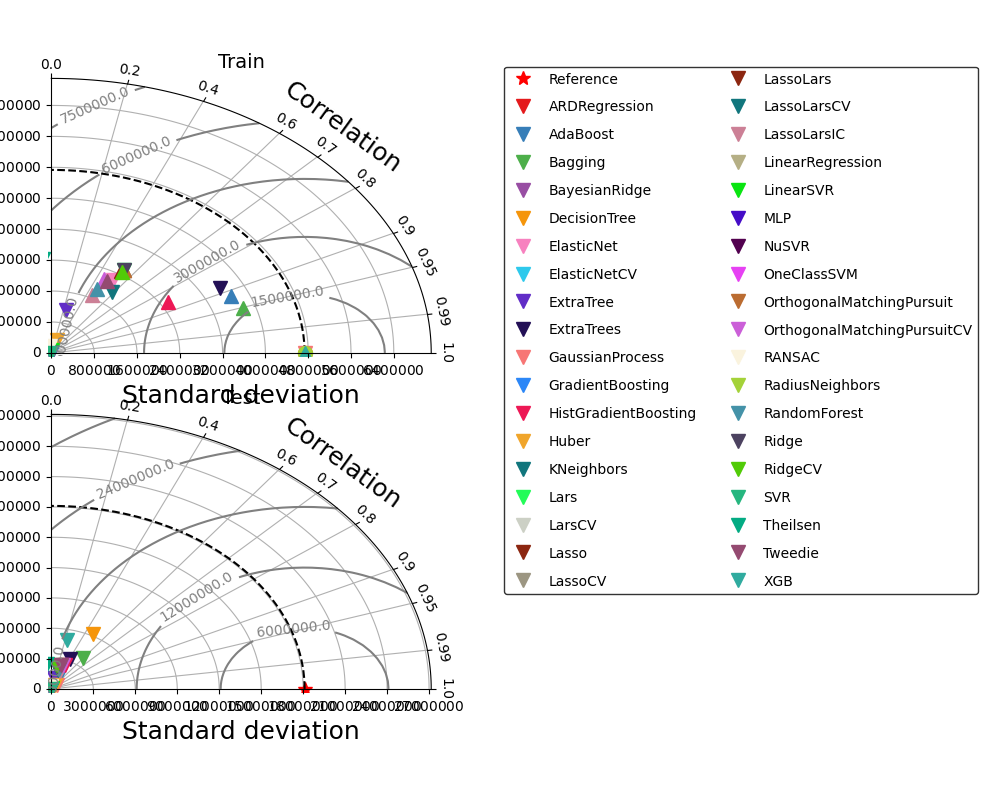
[12]:
_ = comparisons.compare_edf_plots(
data=data,
exclude=["DummyRegressor", "XGBRFRegressor",
"SGDRegressor", "KernelRidge", "PoissonRegressor"]
)
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\neighbors\_regression.py:482: UserWarning: One or more samples have no neighbors within specified radius; predicting NaN.
warnings.warn(empty_warning_msg)
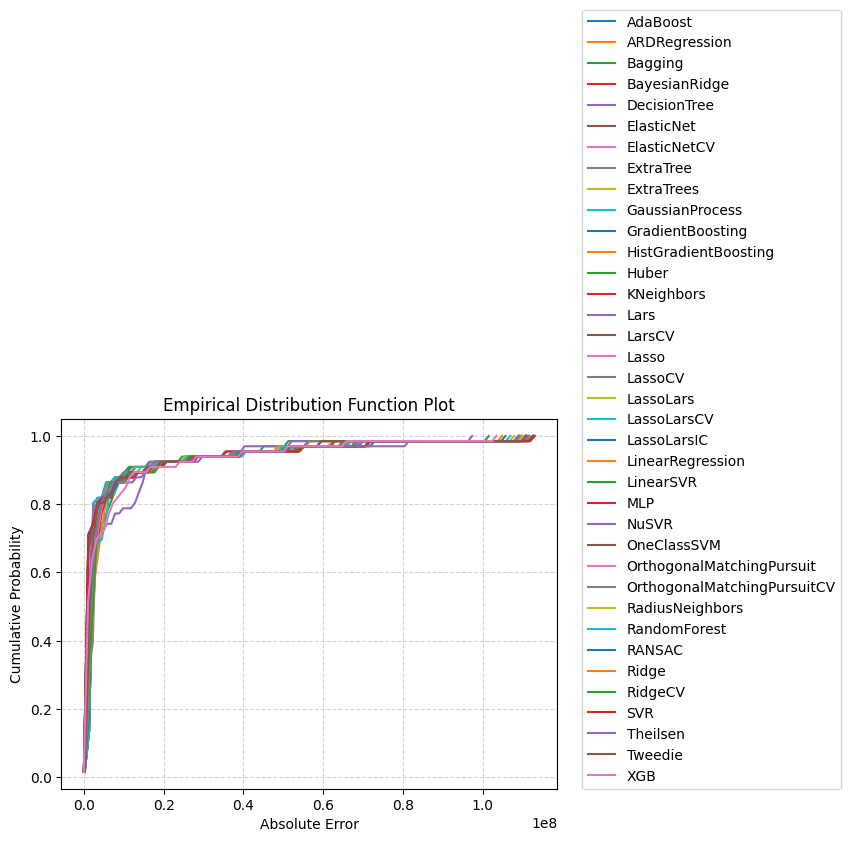
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\neighbors\_regression.py:482: UserWarning: One or more samples have no neighbors within specified radius; predicting NaN.
warnings.warn(empty_warning_msg)
Model RadiusNeighborsRegressor only predicted nans
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\neighbors\_regression.py:482: UserWarning: One or more samples have no neighbors within specified radius; predicting NaN.
warnings.warn(empty_warning_msg)