[ ]:
%matplotlib inline
Comparison of XGBRegressor with different transformations
[1]:
from ai4water.datasets import busan_beach
from ai4water.utils.utils import get_version_info
from ai4water.experiments import TransformationExperiments
from ai4water.hyperopt import Categorical, Integer, Real
from ai4water.utils.utils import dateandtime_now
for k,v in get_version_info().items():
print(f"{k} version: {v}")
python version: 3.8.13 | packaged by conda-forge | (default, Mar 25 2022, 05:59:00) [MSC v.1929 64 bit (AMD64)]
os version: nt
ai4water version: 1.06
tcn version: 3.5.0
xgboost version: 1.7.0
easy_mpl version: 0.21.3
SeqMetrics version: 1.3.3
tensorflow version: 2.7.0
keras.api._v2.keras version: 2.7.0
numpy version: 1.22.2
pandas version: 1.5.1
matplotlib version: 3.6.1
h5py version: 3.6.0
sklearn version: 1.1.3
xarray version: 0.21.1
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\sklearn\experimental\enable_hist_gradient_boosting.py:16: UserWarning: Since version 1.0, it is not needed to import enable_hist_gradient_boosting anymore. HistGradientBoostingClassifier and HistGradientBoostingRegressor are now stable and can be normally imported from sklearn.ensemble.
warnings.warn(
[2]:
data = busan_beach()
input_features = data.columns.tolist()[0:-1]
output_features = data.columns.tolist()[-1:]
[3]:
class MyTransformationExperiments(TransformationExperiments):
def update_paras(self, **kwargs):
y_transformation = kwargs.pop('y_transformation')
if y_transformation == "none":
y_transformation = None
return {
'model': {"XGBRegressor": kwargs},
'y_transformation': y_transformation
}
[4]:
cases = {
'model_None': {'y_transformation': 'none'},
'model_minmax': {'y_transformation': 'minmax'},
'model_zscore': {'y_transformation': 'zscore'},
'model_center': {'y_transformation': 'center'},
'model_scale': {'y_transformation': 'scale'},
'model_robust': {'y_transformation': 'robust'},
'model_quantile': {'y_transformation': 'quantile'},
'model_box_cox': {'y_transformation': {'method': 'box-cox', 'treat_negatives': True, 'replace_zeros': True}},
'model_yeo-johnson': {'y_transformation': 'yeo-johnson'},
'model_sqrt': {'y_transformation': 'sqrt'},
'model_log': {'y_transformation': {'method':'log', 'treat_negatives': True, 'replace_zeros': True}},
'model_log10': {'y_transformation': {'method':'log10', 'treat_negatives': True, 'replace_zeros': True}},
"model_pareto": {"y_transformation": "pareto"},
"model_vast": {"y_transformation": "vast"},
"model_mmad": {"y_transformation": "mmad"}
}
[5]:
num_samples=10
search_space = [
# maximum number of trees that can be built
Integer(low=10, high=30, name='iterations', num_samples=num_samples),
# Used for reducing the gradient step.
Real(low=0.09, high=0.3, prior='log-uniform', name='learning_rate', num_samples=num_samples),
# Coefficient at the L2 regularization term of the cost function.
Real(low=0.5, high=5.0, name='l2_leaf_reg', num_samples=num_samples),
# arger the value, the smaller the model size.
Real(low=0.1, high=10, name='model_size_reg', num_samples=num_samples),
# percentage of features to use at each split selection, when features are selected over again at random.
Real(low=0.1, high=0.5, name='rsm', num_samples=num_samples),
# number of splits for numerical features
Integer(low=32, high=50, name='border_count', num_samples=num_samples),
# The quantization mode for numerical features. The quantization mode for numerical features.
Categorical(categories=['Median', 'Uniform', 'UniformAndQuantiles',
'MaxLogSum', 'MinEntropy', 'GreedyLogSum'], name='feature_border_type')
]
[6]:
x0 = [10, 0.11, 1.0, 1.0, 0.2, 45, "Uniform"]
[7]:
experiment = MyTransformationExperiments(
cases=cases,
input_features=input_features,
output_features = output_features,
param_space=search_space,
x0=x0,
verbosity=0,
split_random=True,
exp_name = f"xgb_y_exp_{dateandtime_now()}",
save=False
)
[8]:
experiment.fit(data = data, run_type='dry_run')
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
running None model
[21:37:25] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running minmax model
[21:37:28] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running zscore model
[21:37:30] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running center model
[21:37:33] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running scale model
[21:37:35] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running robust model
[21:37:38] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running quantile model
[21:37:40] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
running box_cox model
[21:37:43] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
running yeo-johnson model
[21:37:45] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
running sqrt model
[21:37:48] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
running log model
[21:37:51] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
running log10 model
[21:37:53] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
running pareto model
[21:37:56] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running vast model
[21:37:58] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
running mmad model
[21:38:01] WARNING: C:/buildkite-agent/builds/buildkite-windows-cpu-autoscaling-group-i-0ac76685cf763591d-1/xgboost/xgboost-ci-windows/src/learner.cc:767:
Parameters: { "border_count", "feature_border_type", "iterations", "l2_leaf_reg", "model_size_reg", "rsm" } are not used.
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
D:\C\Anaconda3\envs\ai4w_exp_py38\lib\site-packages\scipy\stats\_stats_py.py:310: RuntimeWarning: invalid value encountered in log
log_a = np.log(a)
[9]:
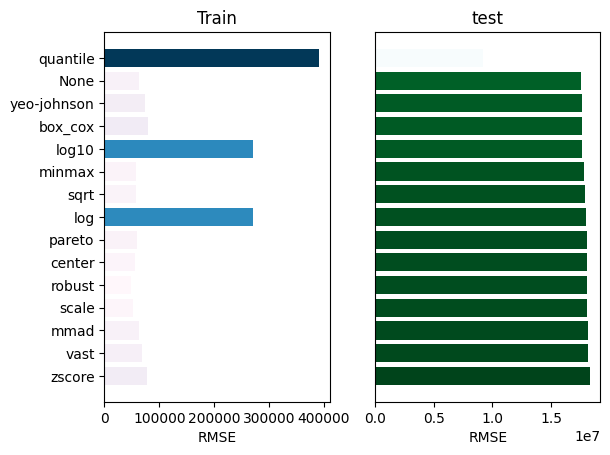
_ = experiment.compare_errors('rmse', data=data)
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
[10]:
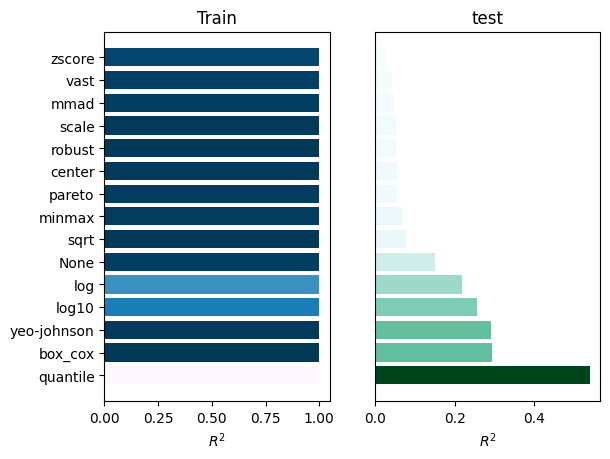
_ = experiment.compare_errors('r2', data=data)
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
[11]:
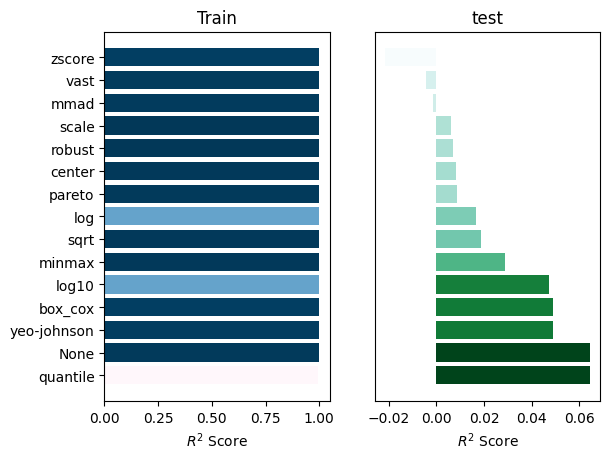
_ = experiment.compare_errors('r2_score', data=data)
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
[12]:
_ = experiment.compare_errors('nrmse', data=data)
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
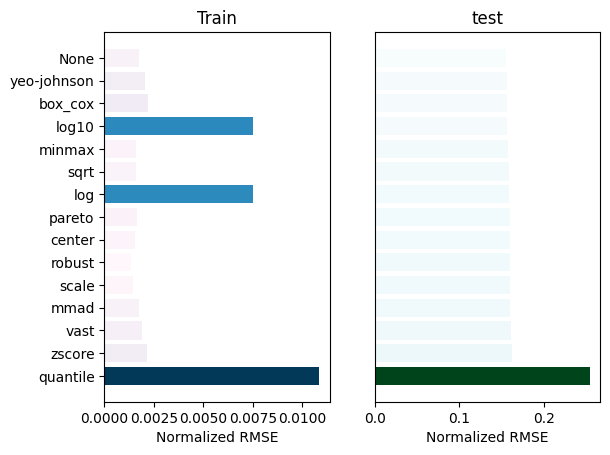
[17]:
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
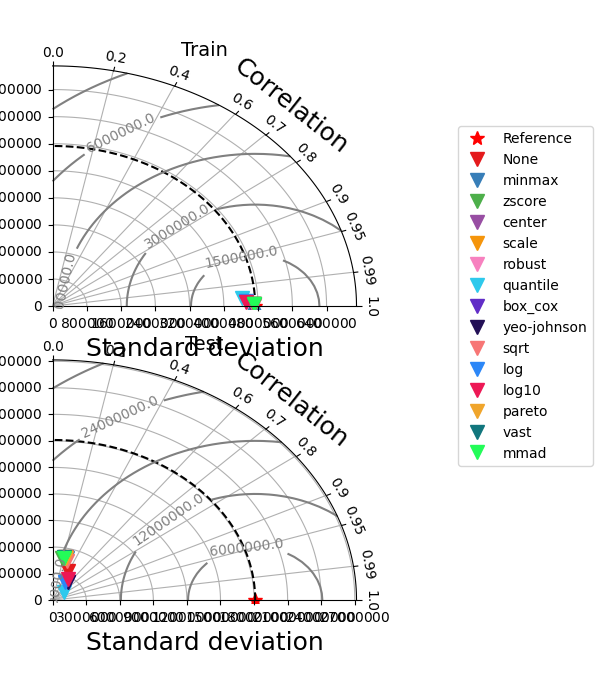
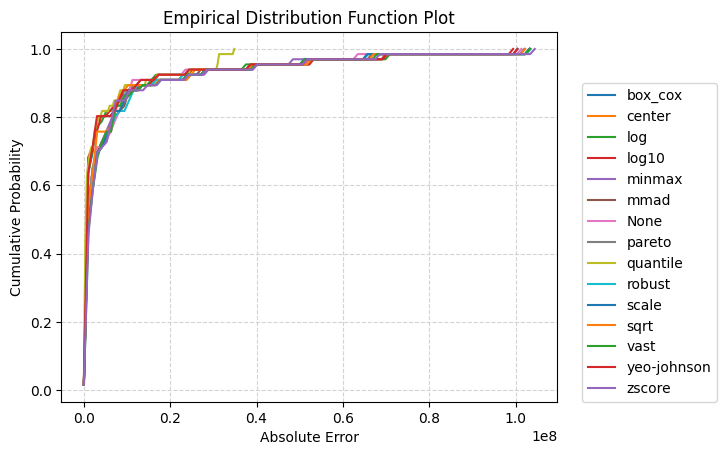
[18]:
_ = experiment.compare_edf_plots(data=data)
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
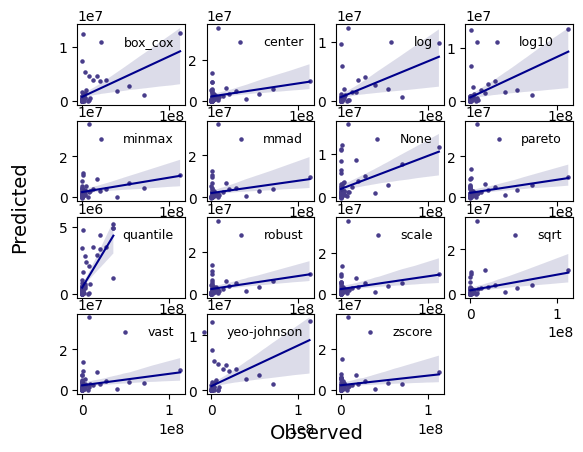
[19]:
_ = experiment.compare_regression_plots(data=data)
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)
[20]:
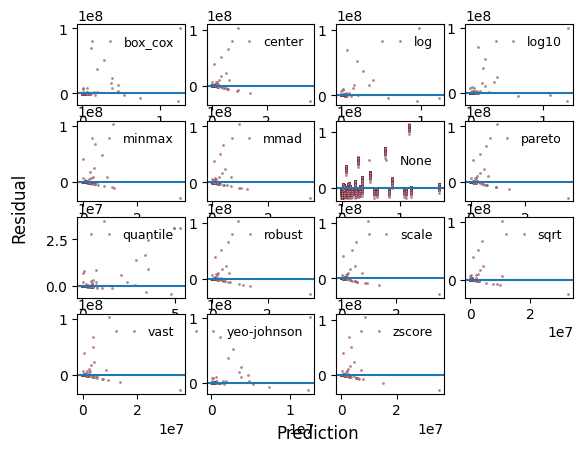
_ = experiment.compare_residual_plots(data=data)
********** Removing Examples with nan in labels **********
***** Training *****
input_x shape: (121, 13)
target shape: (121, 1)
********** Removing Examples with nan in labels **********
***** Validation *****
input_x shape: (31, 13)
target shape: (31, 1)
********** Removing Examples with nan in labels **********
***** Test *****
input_x shape: (66, 13)
target shape: (66, 1)